FAQ¶
Where can I get the stand-alone version of RBPBench?
For the Linux command line version of RBPBench on which the webserver version is based on, please visit the RBPBench GitHub repository. There you can also detailed installation instructions, as well as usage examples and a comprehensive documentation.
What organisms are supported?
Currently the database contains exclusively human RBP binding motifs. However, RBPBench is not restricted to human datasets, as the user can supply any genome file and define a custom motif database, or add additional motifs to search with the present database motifs. If you have motifs that you would like to have included, feel free to contact us (e.g., by opening up a new issue on GitHub).
Where to send bug reports and feature requests?
You can either open up a new issue in the RBPBench GitHub repository (especially for feature requests and command line related bugs), or send bug reports directly on Galaxy in case of faulty job runs.
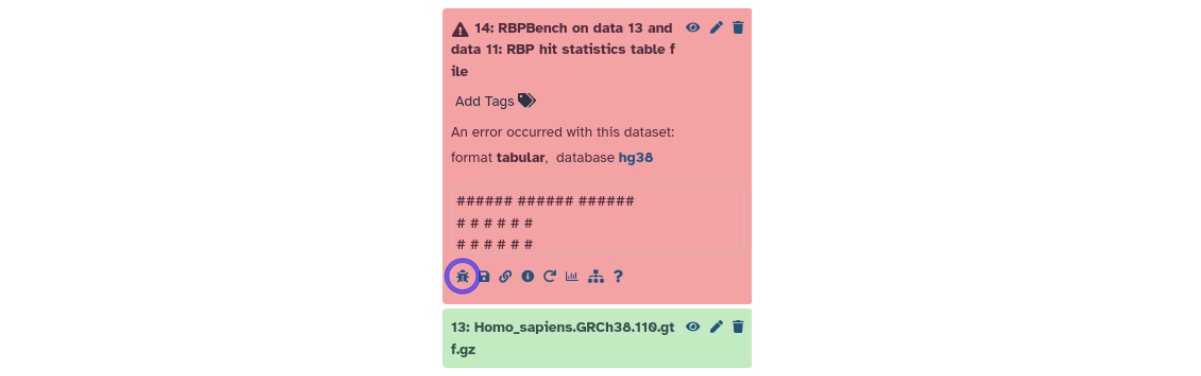 Figure: History output files in red signal jobs with errors.
Click the bug icon (marked by violet circle) to generate a bug report.
Figure: History output files in red signal jobs with errors.
Click the bug icon (marked by violet circle) to generate a bug report.
