Welcome to RBPBench!¶
RBPBench is multi-function tool to evaluate CLIP-seq and other genomic region data using a comprehensive collection of known RNA-binding protein (RBP) binding motifs. RBPBench can be used for a variety of purposes, from RBP motif search (database or user-supplied RBPs) in genomic regions, over motif co-occurrence analysis, to benchmarking CLIP-seq peak caller methods as well as comparisons across cell types and CLIP-seq protocols.
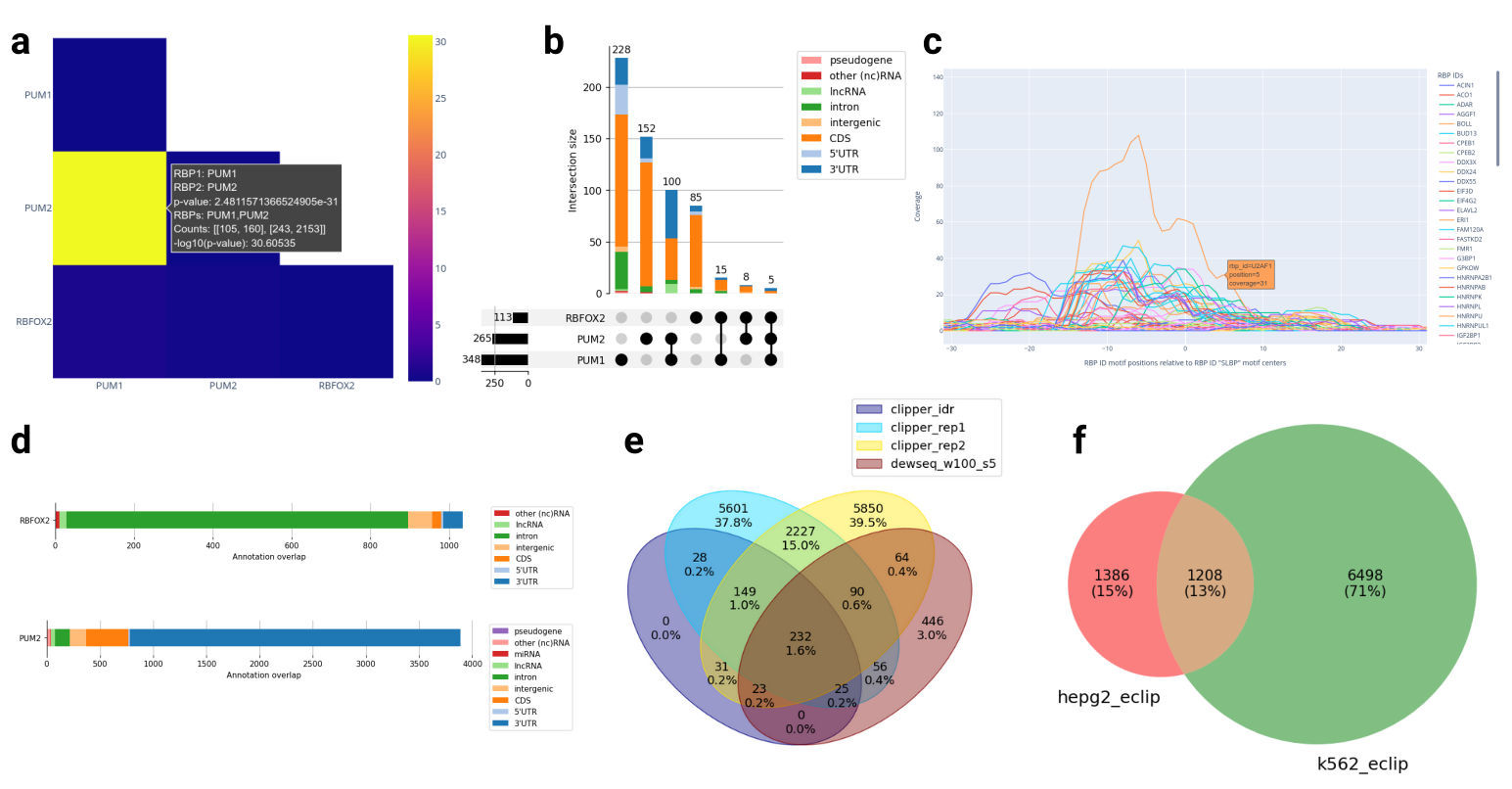 Figure: Some example visualizations produced by RBPBench.
a: RBP binding motif co-occurrence heat map.
b: What RBP binding motif combinations appear in the data.
c: Motif distances in the data relative to an RBP of interest.
d: To which region types RBPs preferentially bind to.
e: Comparing CLIP-seq peak calling methods.
f: Comparing experimental conditions.
Figure: Some example visualizations produced by RBPBench.
a: RBP binding motif co-occurrence heat map.
b: What RBP binding motif combinations appear in the data.
c: Motif distances in the data relative to an RBP of interest.
d: To which region types RBPs preferentially bind to.
e: Comparing CLIP-seq peak calling methods.
f: Comparing experimental conditions.
